Research
Genome folding and the epi-unknome
While we often speak informally of genome folding, it's important to remember that genomes are not really "folded" in the well-defined native structures that protein subunits often are (and, well, many proteins aren't either!). Instead it's better to think of chromosomes as long, disordered chromatin fibers being coerced by different organizing processes. Recent research spurred by innovations in Chromosome Conformation Capture (3C) technologies have revealed two major processes shaping chromosome organization in vertebrates: loop extrusion and compartmentalization [1,2].
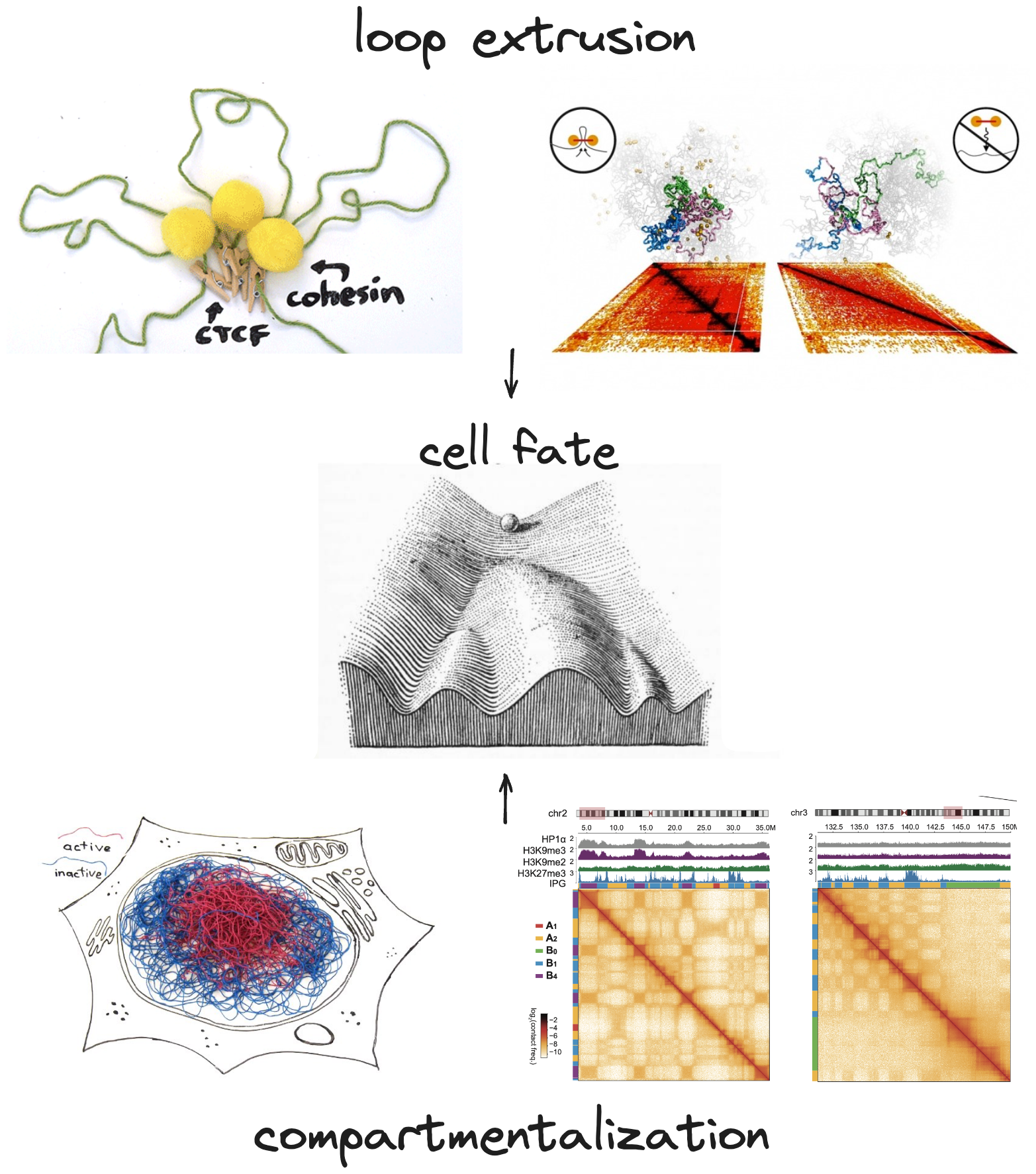
These processes are thought to be critical for various functions in chromosome homeostasis and cell cycle progression, but also in epigenetic regulation and gene regulatory communication, and thus are critical for cell decision making during development and can go awry in aging and disease. The rich data we can gather about these organizing processes using modern genomic technologies also gives us a unique window to discover and decipher new epigenetic states and biophysical mechanisms and elucidate their roles in health and disease [3]. We work on these problems as part of large collaborative efforts in the NIH 4D Nucleome Consortium.

Multi-omics for health and disease
We proud to be members of the newly established NIH Multi-Omics for Health and Disease (MOHD) consortium. The MOHD consortium will work to validate and enhance generalizable multi-omics approaches to identify meaningful biological changes related to health and disease in ancestrally diverse populations, through longitudinal studies from six disease study centers across the country.

As part of MOHD's Data Analysis and Coordination Center (DACC), we will contribute to developing generalizable data harmonization, integration, visualization and analysis methods, best practices, and standards for this unique multidimensional dataset as a resource for the research community.
Foundational software infra for genomic data science
Because of the specialized nature of genomic and multi-omic data modalities and the enormous volumes of data that keep accumulating, an impressive legacy of "bioinformatic" technologies dominates the way we process, store, analyze, visualize, and model omic information.
However, this technology stack has become largely insulated from the rapidly evolving data science and AI/ML ecosystems. We think that the right foundational and middleware glue is necessary to make genomic data truly findable, accessible, interoperable and reusable (FAIR) with the tools of these broader ecosystems. We are involved in several projects and communities that work towards that goal.
References
- 📄 W Schwarzer*, N Abdennur*, A Goloborodko*, et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551:51-56 (2017). doi: 10.1038/nature24281
- 📄 LA Mirny, M Imakaev, and N Abdennur. Two major mechanisms of chromosome organization. Curr Opin Cell Biol 58:142-152 (2019). doi: 10.1016/j.ceb.2019.05.001
- 📄 G Spracklin*, N Abdennur*, et al. Diverse silent chromatin states modulate genome compartmentalization and loop extrusion barriers. Nat Struct Mol Biol 30:38-51 (2023). doi: 10.1038/s41594-022-00892-7
See a full list of publications here.